Our goal is to understand the general principles of organized brain dynamics:
- How do we describe them: are there conserved statistical structures and spatiotemporal motifs in neural activity?
- Where do they come from: what are the cellular and network mechanisms underlying these different patterns?
- What are they useful for: what functions do they serve in behaviorally relevant computations?
To tackle these questions, we leverage a broad range of computational methods—including deep learning, simulator models, statistical data analysis, and signal processing—to analyze and interpret neural activity recordings and multimodal brain data. In parallel, we also contribute methodological advances and best practices that strengthen the application of AI for Neuroscience (AI4Neuro) research.
In the long-term, we hope to uncover general principles of neural dynamics, as well as to create theoretical and computational tools that can inform translational research and clinical applications.
Research Areas
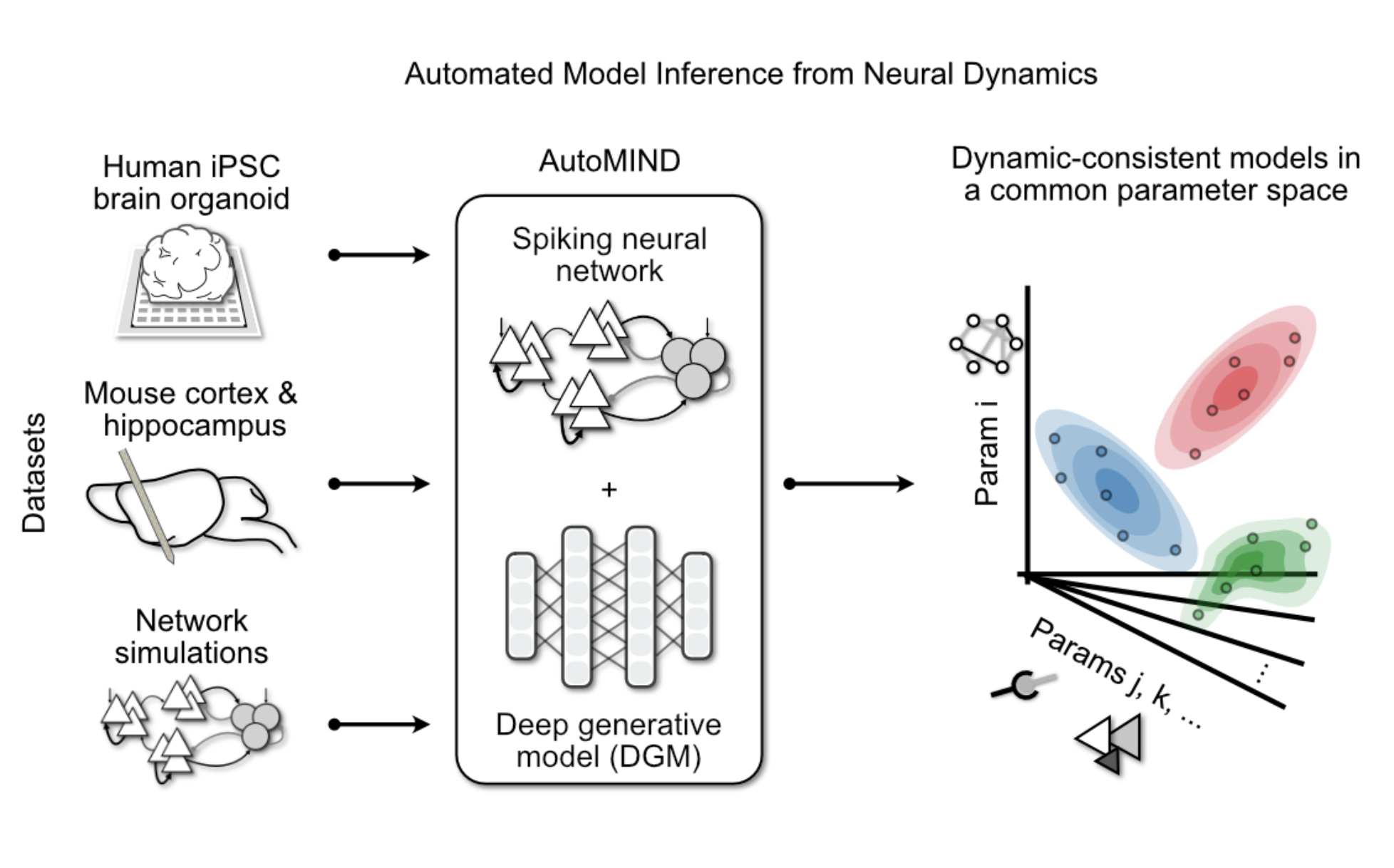
Inverse mechanistic modeling of neural circuit dynamics
We leverage computational frameworks, such as simulation-based inference, that combine probabilistic machine learning and biophysical models to infer circuit mechanisms underlying neural dynamics from brain recordings.
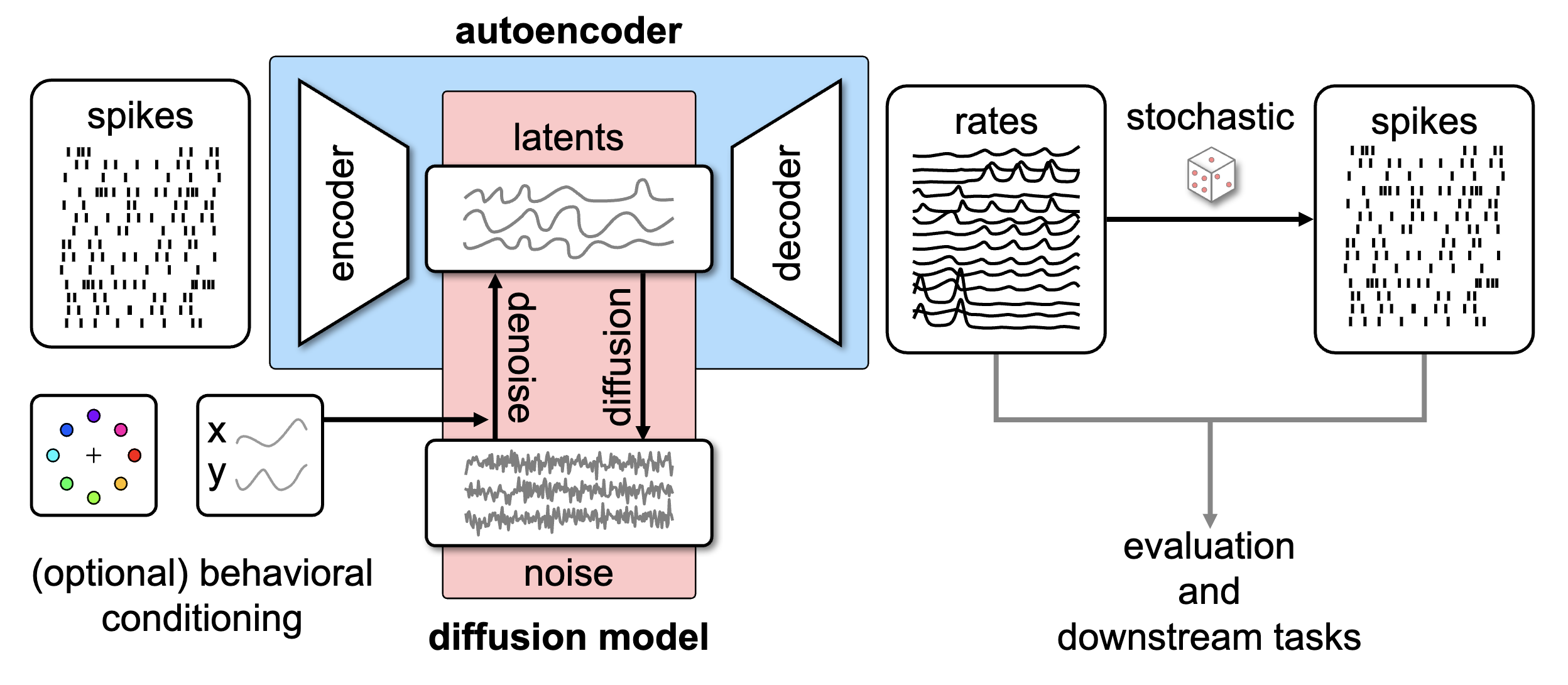
Foundation models of neural time series data
We develop self-supervised learning and generative modeling techniques to extract meaningful representations from large-scale datasets of neural activity recordings, enabling the discovery of latent structures and synthesis realistic neural data.

Neural signal processing and multimodal data integration
We combine data across species, scales, and modalities to derive novel features of neural activity—with a special focus on aperiodic and oscillatory dynamics—and link them to circuit mechanisms and function.